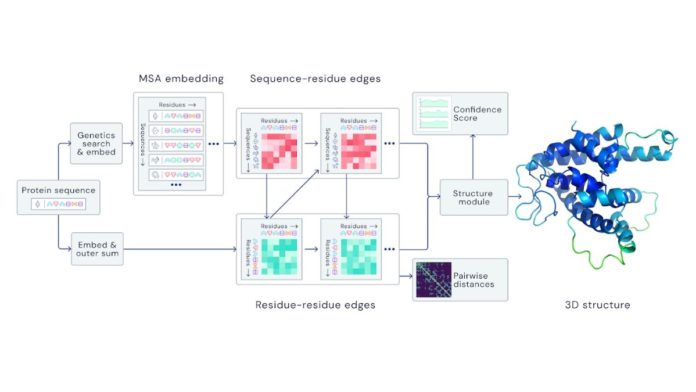
Google’s DeepMind partnered with the European molecular biology laboratory’s European bioinformatics institute (EMBL-EBI) for developing an AI system called ‘AlphaFold’ to predict the three-dimensional structure of a protein by recognizing the sequence of amino acids.
Proteins are extremely complex substances providing structure to cells and organisms. However, they differ from one another primarily by the sequence of amino acids resulting in protein folding.
Protein folding is a physical process where a protein chain arranges to a unique three-dimensional structure. In 1958, Sir John Kendrew and his co-workers came up with a low-resolution 3D structure of the protein. This research had led many scientists to demystify the hidden structures inherited by proteins.
DeepMind tops the list of critical assessments of techniques for protein structure prediction (CASP-14) with high accuracy. The AlphaFold database and source code are freely available to the scientific community, this contribution will aid many advanced biological research.
There is a greater scope in the medical field to understand the viral process and the emerging mutations through a deep learning model for structure prediction. This novel machine learning combines physical and biological knowledge of protein structure to leverage multi-sequence alignments to design a deep learning model.
“This will be one of the most important datasets since the mapping of the Human Genome,” said EMBL Deputy Director-General, and EMBL-EBI Director Ewan Birney. AlphaFold has provided 20,000 proteins of the human genome along with proteomes of 20 other biological organisms summing to 3,50,000 protein structures.
All the data provided is freely available for academic and commercial use under Creative Commons Attribution 4.0 (CC-BY 4.0) license terms. In the coming months, this organization plans to expand the database to cover a large proportion (almost 100 million structures) of all cataloged proteins.